Snake eating Frog

Figure 1. Snake eating a Frog. Snakes are able to unhing their jaws to eat prey that is larger than their heads. Creative common licensed. Image by Brian Gratwicke. https://flic.kr/p/9NtqUM

Figure 1. Snake eating a Frog. Snakes are able to unhing their jaws to eat prey that is larger than their heads. Creative common licensed. Image by Brian Gratwicke. https://flic.kr/p/9NtqUM

Figure 1. Oxalis Triangularis Development. Comparison of the change in pigmententation during development. Immature speciemens (right) produce green foliage. Mature (left) speciments produce purple leaves. Creative commons licensed image by https://flic.kr/p/frB4rf.

Figure 1. The tongue of a giraffe. The giraffe is one of three animals known to have a blue tongue. Creative commons licensed image by Robyn Jay https://flic.kr/p/gthW8a

Figure 1 - The Thylacine, Thylacinus cynocephalus, is an extant species of carnivorous marsupial that was hunted to extinction by farmers because of predation of domestic sheep.
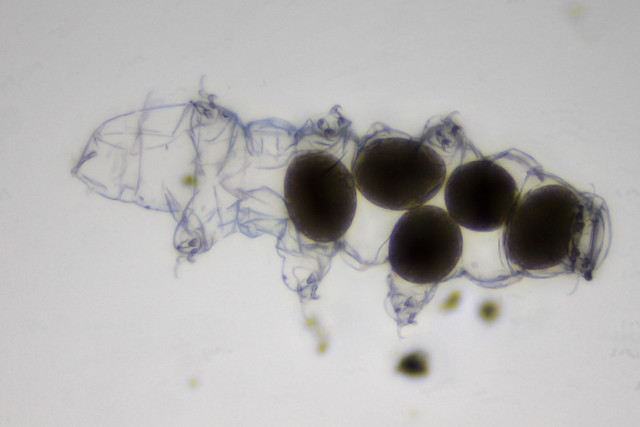
Figure 1. Tardigrade with eggs. Focus stacked image of non-living tardigrade. Creative commons licensed image by Specious Reasons https://flic.kr/p/ZHFAdR
Ethnonationalism is like nationalism in that it is a way to find sense and belonging within a group, but ethnonationalism emphasises the person’s ethnicity rather than where they live. These movements band people together based on their ethnicity regardless of their location. An example of this is seen during the fall of the Soviet Union, which encompassed many satellite countries near Russia such as Latvia, Estonia, and Lithuania. At the time they weren’t countries, but as the citizens began to see the Soviet Union fall, they began to identify with their region and ethnic identities rather than with the union they were a part of. Another example is Israel’s “right to return”, which grants every ethnically Jewish person the right to return to Israel, and be granted citizenship. This shows they identify with a place they may not live.
Protein misfolding can occur through a number of mechanisms and lead to a large variety of disease and misfunction. One pathway for protein misfolding and pathogenesis is improper degradation of proteins. Improper degradation occurs when proteins that are partially functional and can actually benefit cellular processes are degraded despite it being detrimental to the cell. This is seen in the case of cystic fibrosis, where a deletion of a phenylalanine in CFTR leads to partial functionality but is still targeted for degradation by CHIP, a molecular chaperone which ubiquitylates the protein. CFTR is an important membrane channel for the production of mucus, which is why this improper degradation is seen in a large number of cystic fibrosis patients. Another way in which improper folding can lead to disease is through improper localization. Improper localization occurs when misfolded proteins cannot get to where they need to go, leading to not only a loss-of-function but potential toxicity if aggregated in the wrong place. One example of this is misfolded antitrypsin, which becomes retained in the ER of liver cells and accumulates, preventing synthesis of other proteins resulting in liver damage. Also, since antitrypsin does not get secreted to its proper location, it is unable to inhibit protease activity in the lungs leading to damage in the alveoli and emphysema. Another mechanism for pathogenesis as a result of protein misfolding is dominant-negative mutations. Dominant-negative mutations are characterized by mutant proteins that compromise the function of wild-type proteins, most often in a dimer or quaternary structure. An example of this process is seen in the connective tissue disorder epidermolysis bullosa simplex. When mutant forms of keratin proteins are present, they disrupt the function of the entire keratin composed filament, leading to fragile skin that blisters easily in response to minor friction. Gain of toxic function and amyloid accumulation are two other mechanisms for pathogenesis as a result of misfolding and play a big role in neurodegenerative disorders.
FIGURE 1
What are we dealing with? Tip-links connect two stereocilia = mechanical sensitivity
- it is risky to include the model at the beginning because it can lead to circular logic but it can also help us as readers to understand the paper better (image k)
- why use guinea pig because they have lower frequency hearing + longer stereocilia, b/c w/ higher frequency hearing you have a lot shorter stereocilia
- it is easier to identify locations with longer stereocilia
- if you break the tip link, the CDH23 will move up, this is one way to prove that the cadherin is part of the TL
- they can predict the position of each of the antibodies from the base of the lower stereocilia
- b/c you know the sequence and repetition, you can predict the distance b/w the EC domains
- the experimental results are somewhat far from the expected distances for images f-h, is this good enough? yes
- you are req’d to provide a SD, you can do this by providing a bar graph with SD error bars (for images f-h) but it is better to
- there are 40 measurements falling between 1-20 nm and if you average all of them together get 37 for instance
FIGURE 2
- His tag used to purify the protein,
- there is a homodimer and multiple strands to anchor into the membrane
- there is better agreement
- why is there such a small difference compared to the experimental results they show in Figure 1
- it is in a much controlled environment, you already expressed the protein, and purified it
- image 1 is in the live cell and can have different tension b/c of position in stereocilia which gives them different lengths
- what about orientation, it looks like 100% have parallel arrangement of filaments, homodimers
- the evidence looks very convincing but of the 195, only 131 were on one end
On the methods project -
This morning I went to the greenhouses that we have here at the university. The greenhouse I entered was called the “tropical garden” I believe. This is something that I will definitely have to double check. The species name of the flower I chose is: Haemaria discolor. If belongs to the genus: Ludisia and to the family or: Orchidaceae. It’s common name is the Jewel Orchid. It appeared to be in its optimal environment because it was in full bloom, looked very “plump” and healthy. It was definetly a beautiful flower and caught my eye because it closely resembles the flowers that grew near my childhood home (their species name is: Convallaria majalis, but most people know them as the Lily of the Valley). Another intriguing characteristic of these flowers were their leaves. They are about 3 inches long and are a deep green color with some hues of maroon. They also have white lines that run parallel to each other from the base of the leaf towards the peak. Their stem is long and thin and the flowers that have already budded are white with some type of yellow center.
Recent comments