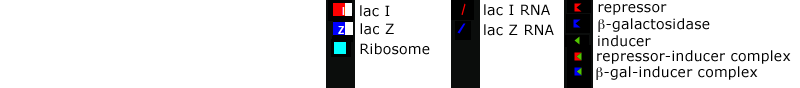
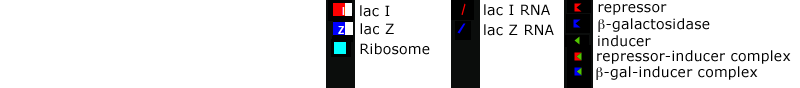
created with NetLogo
view/download model file: lac_operon.nlogo
WHAT IS IT?
-----------
This is a model of the regulation of gene expression based on the lac operon: a bacterial gene that produces an enzyme that breaks down lactose, a complex sugar, into simpler sugars: glucose and galactose. This gene is only transcribed when lactose is in the environment. This system was one of the first models of gene expression described.
HOW IT WORKS
------------
In this system, there are two genes: the lacI gene produces a repressor molecule which in turn binds to the operon of the lacZ gene which codes for beta-galactosidase, an enzyme. As long as the repressor is bound to the operon, the beta-galactosidase gene is turned off.
When a gene is active, RNAs (represented by circles) are produced and diffuse to ribosomes (represented by squares) where they are translated into proteins. When an inducer is added the system, it binds to the repressor molecules and prevents them from binding to the lacZ operon. This causes the lacZ gene to begin transcribing RNAs which are translated into beta-galactosidase. The beta-galactosidase molecules bind to and cleave the inducer molecules. When inducer molecules are no longer available, the repressor molecules bind to the operon again, shutting the system off.
This implementation of the model makes a variety of assumptions. First, the model assumes that proteins are degraded by the cell over time. The number of proteins at any one time are the result of a balance between new proteins being added and old ones being removed through degradation.
In real systems, the coding region for lacZ is followed by lacY and lacA, which code for other proteins and are transcribed at the same time as lacZ. These other proteins do not contribute directly to the regulatory control of lacZ and have not been included in the model for simplicity.
HOW TO USE IT
-------------
Click "setup" to create the genes and ribosomes.
Click "go" to start the model.
Switches allow the user to add inducer and select the number of molecules to add per unit time.
Other switches allow turning off (or "knocking out") the lacI and lacZ genes.
Two plots: one showing the current concentration of beta-galactosidase and the other showing levels of repressor, beta-galactosidase, and their complexes with inducer molecules.
THINGS TO NOTICE
----------------
There are a few repressor molecules when the model first turns on (as long as the lacI gene hasn't been turned off). Note that very quickly after the model starts, one of the repressors has bound to the lacZ operon.
Every now and then, you'll see the lacZ gene produce RNA even when the repressors are available. What hypotheses can you propose for why this might happen?
THINGS TO TRY
-------------
Predict at which level you believe the lacZ gene will begin operating and then try adding various levels of inducer. What differences do you notice between different levels?
Predict the behavior of the system when you turn off each of the genes and then test your predictions.
When the lacZ gene switches on, it produces a distinctive curve in the levels of concentration of beta-galactosidase. What is happening at each stage of the curve? What happens when the gene switches off.
EXTENDING THE MODEL
-------------------
To simplify this model, a number of factors have been de-paramaterized, including the rates of each gene (dI and dZ) and the coefficients of binding and dissociation (KB and Kd). By adding sliders, you could extend this model and explore the behavior of the system with different values.
CREDITS AND REFERENCES
----------------------
This model was created as part of the 2004 BioQUEST Curriculum Consortium
Summer Workshop.
Copyright 2004 by Steven Brewer, Pat Ehrman, and Allen Koop. All rights reserved.
This model was inspired by many of the sample Netlogo models and parts were based on functions from the Enzyme Kinetics model. (Wilensky, U. (2001). NetLogo Enzyme Kinetics model. http://ccl.northwestern.edu/netlogo/models/EnzymeKinetics. Center for Connected Learning and Computer-Based Modeling, Northwestern University, Evanston, IL.)
Permission to use, modify or redistribute this model is hereby granted,
provided that both of the following requirements are followed:
a) this copyright notice is included.
b) this model will not be redistributed for profit without permission
from the authors.
Contact the authors for appropriate licenses for redistribution for
profit.
To refer to this model in academic publications, please use:
Brewer, S.D. Ehrman, P., and Koop, A. (2004). A Netlogo Model of lac operon Gene Expression. http://bcrc.bio.umass.edu/netlogo/models/LacOperon
Biology Computer Resource Center
In other publications, please use:
Copyright 2004 by Steven Brewer, Pat Ehrman, and Allen Koop. All rights reserved. See
http://bcrc.bio.umass.edu/netlogo/models/LacOperon
for terms of use.